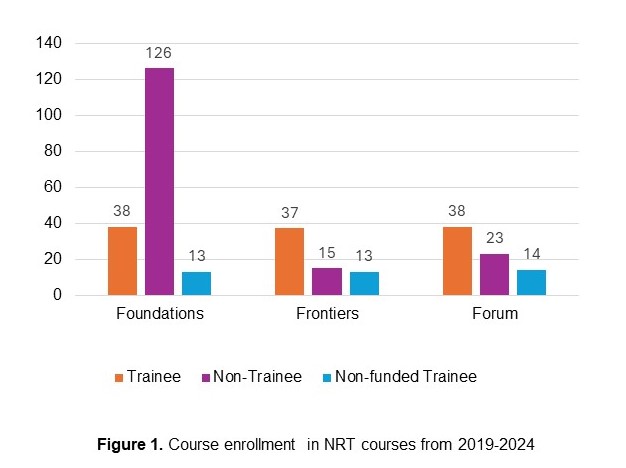
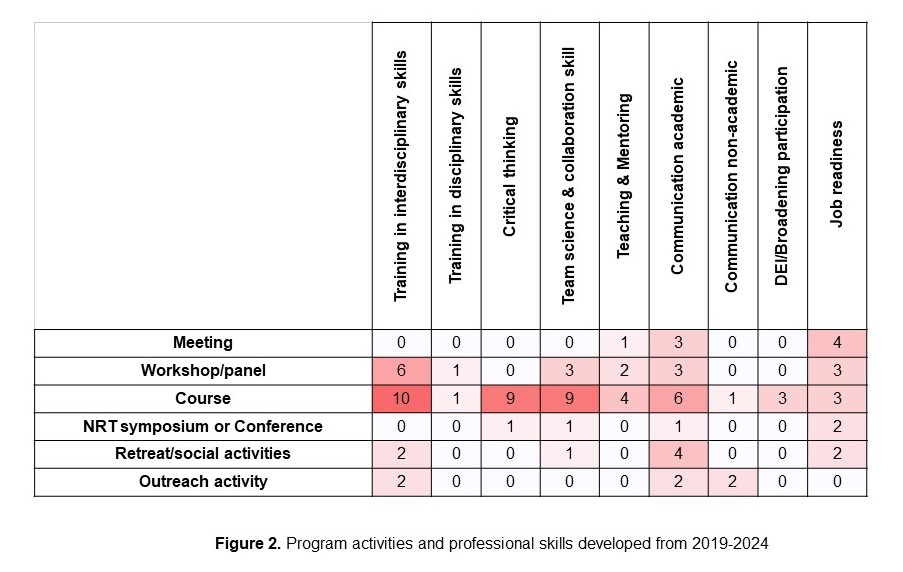
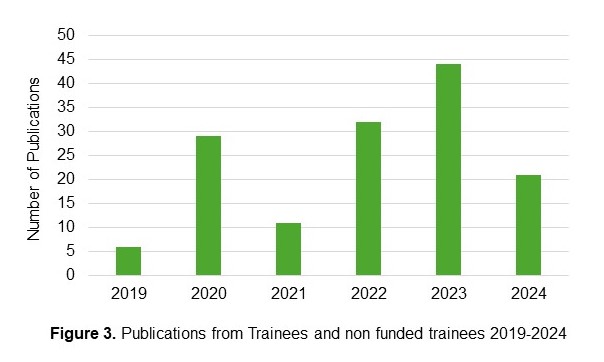
Products
Outcomes
Publications:
2022-23
Palande, S., Kaste, J.A., Roberts, M.D., Segura Aba, K., Claucherty, C., Dacon, J., Doko, R., Jayakody, T.B., Jeffery, H.R., Kelly, N. and Manousidaki, A., 2022. The topological shape of gene expression across the evolution of flowering plants. bioRxiv, pp.2022-09.
Doko, R. and Liu, K.J., 2023. The Impact of Multiple Sequence Alignment Error on Phylogenetic Estimation under Variable-Across-Phylogeny Substitution Models. bioRxiv, pp.2023-01.
MacCready, J.S., Roggenkamp, E.M., Gdanetz, K. and Chilvers, M.I., 2023. Molecular Plant-Microbe Interactions.
Zhang, C., Johnson, N., Hall, N., Tian, X., Yu, Q. and Patterson, E., 2023. Subtelomeric 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) copy number variation confers glyphosate resistance in Eleusine indica.
Bryson, A.E., Lanier, E.R., Lau, K.H., Hamilton, J.P., Vaillancourt, B., Mathieu, D., Yocca, A.E., Miller, G.P., Edger, P.P., Buell, C.R. and Hamberger, B., 2023. Uncovering a miltiradiene biosynthetic gene cluster in the Lamiaceae reveals a dynamic evolutionary trajectory. Nature Communications, 14(1), p.343.
2021-22
Kaste, J.A., Green, A. and Shachar-Hill, Y., 2022. Integrative Teaching of Metabolic Modeling and Flux Analysis with Interactive Python Modules. bioRxiv, pp.2022-11.
Kaste, J.A. and Shachar-Hill, Y., 2022. Accurate flux predictions using tissue-specific gene expression in plant metabolic modeling. bioRxiv, pp.2022-09.
Ford, K.C., Kaste, J.A., Shachar-Hill, Y. and TerAvest, M.A., 2022. Flux-Balance Analysis and Mobile CRISPRi-Guided Deletion of a Conditionally Essential Gene in Shewanella oneidensis MR-1. ACS Synthetic Biology.
Ranaweera, T., Brown, B.N., Wang, P. and Shiu, S.H., 2022. Temporal regulation of cold transcriptional response in switchgrass. bioRxiv.
Wang, P., Schumacher, A.M. and Shiu, S.H., 2022. Computational prediction of plant metabolic pathways. Current Opinion in Plant Biology, 66, p.102171.
Bird, K. A., Jacobs, M., Sebolt, A., Rhoades, K., Alger, E. I., Colle, M., ... & Edger, P. P. (2022). Parental origins of the cultivated tetraploid sour cherry (Prunus cerasus L.). Plants, People, Planet.
Teresi, S. J., Teresi, M. B., & Edger, P. P. (2022). TE Density: a tool to investigate the biology
of transposable elements. Mobile DNA, 13(1), 1-18.
Xu, Y., Wieloch, T., Kaste, J. A. M., Shachar-Hill, Y., & Sharkey, T. D. (2022). Reimport of carbon from cytosolic and
vacuolar sugar pools into the Calvin–Benson cycle explains photosynthesis labeling
anomalies. Proceedings of the National Academy of Sciences, 119(11).
Wilson, M. L., & VanBuren, R. (2022). Leveraging millets for developing climate resilient agriculture. Current
Opinion in Biotechnology, 75, 102683.
Hoopes, G., Meng, X., Hamilton, J. P., Achakkagari, S. R., Guesdes, F. D. A. F., Bolger,
M. E., Bornowski, N.,... & Finkers, R. (2022). Phased, chromosome-scale genome assemblies of tetraploid
potato reveals a complex genome, transcriptome, and predicted proteome landscape underpinning
genetic diversity. Molecular Plant.
Sadohara, R., Izquierdo, P., Couto Alves, F., Porch, T., Beaver, J., Urrea, C. A., & Cichy, K. (2022). The Phaseolus
vulgaris L. Yellow Bean Collection: genetic diversity and characterization for cooking
time. Genetic Resources and Crop Evolution, 1-22.
2020-21
Sadohara, R., Long, Y., Izquierdo, P., Urrea, C. A., Morris, D., & Cichy, K. (2021). Seed coat color genetics and genotype× environment effects in yellow beans via machineâ€learning and genomeâ€wide association. The Plant Genome, e20173.
Bryson, A.E., Wilson Brown, M., Mullins, J., Dong, W., Bahmani, K., Bornowski, N., Chiu, C., Engelgau, P., Gettings, B., Gomezcano, F. and Gregory, L.M., 2020. Composite modeling of leaf shape along shoots discriminates Vitis species better than individual leaves. Applications in plant sciences, 8(12), p.e11404.
Zhou, P., Li, Z., Magnusson, E., Gomez Cano, F., Crisp, P.A., Noshay, J.M., Grotewold, E., Hirsch, C.N., Briggs, S.P. and Springer, N.M., 2020. Meta gene regulatory networks
in maize highlight functionally relevant regulatory interactions. The Plant Cell, 32(5), pp.1377-1396.
Dale, R., Oswald, S., Jalihal, A., Laporte, M.F., Fletcher, D.M., Hubbard, A., Shiu, S.H., Nelson, A.D. and Bucksch, A., 2021. Overcoming the challenges to enhancing experimental
plant biology with computational modeling.
Educational Materials:
Phenology data visualization tool
Mini Data Carpentry Genomics Workshop
Lightning talk rubric
Imposter syndrome poster and presentation